Storing Digital Information in DNA – a Solution for the Future?
 [1]Throughout history, information has been stored through ever-changing channels, and increasingly people store pictures, work and other important information digitally. But how do we ensure that important information can be preserved for the long-haul, for even tens of thousands or hundreds of thousands of years? Presenting at the American Chemical Society (ACS) Conference and Expo in Boston this week, Robert N. Grass, Ph.D., ETH Zurich, suggests we look at something produced by nature — that’s been around longer than most and has proven to be stable over long durations: DNA.
[1]Throughout history, information has been stored through ever-changing channels, and increasingly people store pictures, work and other important information digitally. But how do we ensure that important information can be preserved for the long-haul, for even tens of thousands or hundreds of thousands of years? Presenting at the American Chemical Society (ACS) Conference and Expo in Boston this week, Robert N. Grass, Ph.D., ETH Zurich, suggests we look at something produced by nature — that’s been around longer than most and has proven to be stable over long durations: DNA.
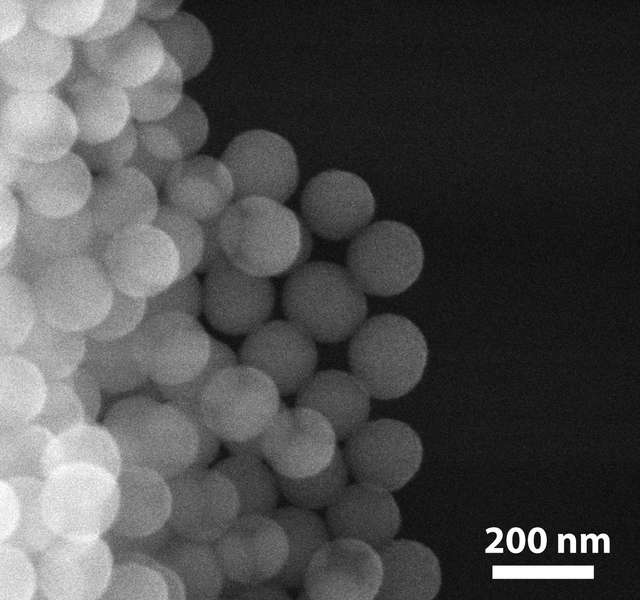
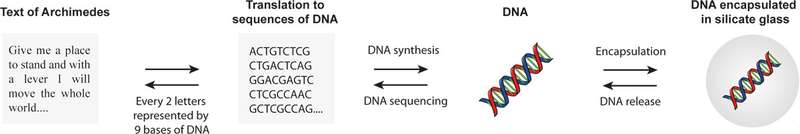
Storing information in DNA could be a way to store large amounts of data in a very stable way, Grass said at an ACS press conference Monday. A paperback book-sized hard drive can back up five terabytes of information, compared to DNA which, in theory, could store more than 300,000 terabytes in a fraction of an ounce. There were two parts – figuring out how to translate the information so it could be read in DNA, and how to physically store the DNA molecule so that it is as stable as a fossil that encodes genomic data for hundreds of thousands of years. The first part is done by translating digital data, that is read by 1s and 0s into DNA base pairs, or A, T, C, and G. To address the second Grass said they “generated an artificial fossil, where the DNA is encapsulated in inorganic material where it’s protected from water, oxygen, and it doesn’t degrade as fast.” The material Grass used is silicon and glass. Grass said glass is used because it can be easily synthesized in water at room temperature, it’s an extremely inert material, and it’s extremely resilient to the environment.
“We use small glass particles that are only a few nanometers large, in which we chemically entrap DNA molecules. In that entrapped state they are almost as stable as they are when they are entrapped in fossil bones of a few hundred thousand year old fossils, which are known to be stable over that time-frame.”
Grass’ team figured out a way to correct for potential mutations when reading what is encoded in the DNA. To test it, the researchers encoded DNA with 83 kilobytes of text from the Swiss Federal Charter from 1291 and the Method of Archimedes from the 10th century. Once the DNA was entrapped in the inorganic spheres, it underwent a heating process involving being warmed to nearly 160 degrees Fahrenheit for one week, which is comparable to keeping it for 2,000 years at 50 degrees. When it was decoded it was error free.
 [2]One large constraint for this type of information storage is expense. Grass said that at the moment there are only a few companies that do DNA synthesis, so the market is very small. This could be remedied if there were a greater need, like what happened in the development of writing DNA, as sequencing technologies are moving rapidly.
[2]One large constraint for this type of information storage is expense. Grass said that at the moment there are only a few companies that do DNA synthesis, so the market is very small. This could be remedied if there were a greater need, like what happened in the development of writing DNA, as sequencing technologies are moving rapidly.
The next step in research, aside from scaling up, is creating a sort of ‘filing system’ where you could read back particular pieces without having to read all of the information contained on the DNA strands.
“In DNA storage, you have a drop of liquid containing floating molecules encoded with information,” Grass said. “Right now, we can read everything that’s in that drop. But I can’t point to a specific place within the drop and read only one file.”
The team is also trying to further improve the stability of the molecule and further understand its decay characteristics.
Grass wants people to think about something that sometimes gets lost in our data-driven world – preserving important information that we have today for future generations.