The Importance of Primer Validation in Real-time PCR and New Tools That Make It Easier
LISTED UNDER:
It’s a tale as old as time. A bride is cooking a roast for her new husband, cutting off the ends the way her mother always did. Amused, the husband asks why she is cutting off the best part. “That’s the way my mother always made it,” she answers. Eager to find out why they both cooked it this way, mother and daughter consult the grandmother. What they discover is that the recipe had only been altered to accommodate a roasting pan that was too small, and for generations they had followed the tradition without really understanding it – in turn, wasting something delicious.
What, you might ask, does story this have to do with research? The lesson in this story is that we all do things a certain way, perhaps without knowing why we do them. The same idea applies in the lab. Researchers may follow real-time PCR (qPCR) “recipes” without necessarily giving them any thought, oftentimes bypassing one of the most important steps: validating their primers.
Real-time quantitative polymerase chain reaction is one of the most ubiquitous laboratory techniques in modern life sciences. It is commonly used for gene expression analyses, diagnosing infectious diseases, detecting food pathogens and more.
Despite being more than two decades old, many researchers continue to perform qPCR incorrectly and overlook the important step of primer validation and optimization. Though in the short-term it can be labor and time intensive, it ensures researchers are only amplifying the stretch of DNA they intend to analyze.
But why exactly is primer validation and optimization so significant?
A tedious task, but a necessary evil
Mark Kibschull, Ph.D., a researcher at Mount Sinai Hospital in Toronto, says that primer validation is one of the most important steps in qPCR. He works with human embryonic and mouse trophoblast stem cells, studying placenta development. Until recently, Kibschull designed his own primers, which he describes as a tedious task with many pitfalls. For every new target, he would create and buy two sets of primer pairs using free software from the National Center for Biotechnology Information (NCBI) website.
Primers by standard software might only detect a specific splice variant, so detailed analysis of the gene of interest is necessary. Kibschull says this is especially important when members of a gene family are characterized. In addition, primers should span an exon-exon junction to prevent gDNA amplification.
When titrated, the self-designed primers showed varying efficiency, often significantly short of the 100 percent (+/- 10 percent) efficiency Kibschull aimed for, so primers with low or “odd” efficiencies would typically be thrown out. In any case, expression analysis has to be corrected for specific primer efficiency within the software, as careful estimation of primer efficiency is necessary to avoid significant measurement inaccuracies.
Another pitfall with self-designed primers was additional peaks on the titration curve, indicating the formation of primer dimers. Kibschull says that every fourth target, he had to re-order primers and that it would take several weeks to find the right pair for these problem targets. Ultimately, he decided, self-designed primers were a waste of his lab’s money and time.
Meredith Tennis, Ph.D., a researcher at the University of Colorado Cancer Center, had a different experience, but one that led to the same conclusion: Validation is a necessary evil. When she first started working with reverse transcription qPCR (RT-qPCR) for lung cancer chemotherapy prevention studies, she and her team would pull primer sequences straight from other researchers’ published papers, assuming they had performed the necessary validation work.
More often than not, though, this wasn’t the case. Tennis was stuck with a lot of extra work, going through lengthy trial and error to find primers that fit her experiments. It wasn’t until companies introduced her to primer validation that Tennis realized a much greater level of qPCR integrity existed. Those companies also began to explain the step ahead of time, making sure researchers understood the value it had on their qPCR experiments.
A lack of efficiency and uniformity
Both Kibschull and Tennis encountered problems, whether through designing their own primers or relying on others’ work. These experiences not only confirmed to both scientists that primer validation is an important step in qPCR experiments, but it is also one that lacks an efficient and uniform protocol. Flaws in this step of the study design can lead to erroneous data. However, because real-time PCR almost always generates results, you may never catch this mistake unless you’re looking for it.
For example, Kibschull says that oftentimes researchers export Cq values from their qPCR system without looking at the data and graphs, mostly ignoring threshold values as the program sets a common threshold for all wells on a plate. In this case, well groups need to be defined when primers have different efficiencies.
Using web-based tools to “validate” their primers, a convenient shortcut for some scientists, can be dangerous. Kibschull says researchers use websites thinking they will deliver exactly what they need, simply because the process is automated. In reality, there are pitfalls to designing primers that can’t be caught with current software systems.
Creating awareness in the scientific community
Another recurring problem is the lack of rules related to primers that publications place on authors. Reviewers seldom check primer sequences used in research papers, possibly leading to the circulation of erroneous data around the scientific community.1
Stephen Bustin, professor of molecular medicine at Anglia Ruskin University, says that most researchers choose to be “blissfully unaware” in their approach to qPCR. He says they trust their qPCR results because they are unwilling to consider the ramifications of research that is based on poor science.
In response, a group of concerned researchers, including Bustin, published a paper in 2009 that outlines the information that should be included in a qPCR paper. These guidelines have come to be known as MIQE (Minimum Information for the Publication of Quantitative Real-Time PCR Experiments).2 This paper lists primer validation as one of the important qPCR steps.
Following MIQE guidelines for assay design, validation and optimization increases the reliability of qPCR data. In addition, doing so allows the reviewer or reader to assess the technical quality of the paper. He or she can then focus on the conclusions drawn from the data without second-guessing the quality of the RNA, whether the qPCR assays were specific and efficient and whether appropriate normalization measures were taken.

A complete solution for reliable results
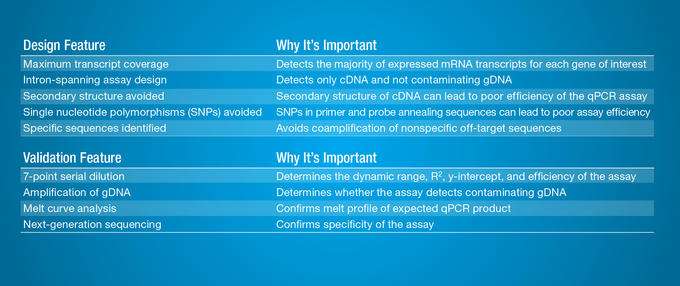
The term “validation” is not well-defined when it comes to qPCR. Many companies offer pre-designed qPCR assays, but the term has many different definitions (Figure 1). To make qPCR even more reliable and introduce a new benchmark in validation beyond industry standards and common research practice, Bio-Rad and qPCR bioinformatics firm Biogazelle designed, optimized and experimentally validated every primer pair for nearly all protein-coding genes in the human, mouse and rat transcriptomes. To ensure quality, every single primer set has been empirically wet-lab validated for specificity, efficiency and sensitivity. To test assay specificity, for instance, Biogazelle scientists validated all PCR products from cDNA using next-generation sequencing, verifying the percentage of on-target amplification. In addition, all of the raw data, except for the primer sequences, is supplied for the customer. The Bio-Rad products take much of the guesswork, time delays and frustration out of the experiment (Figure 2).
PrimePCR products are available as individual SYBR Green and probe-based assays, SYBR Green-based pre-plated pathway- and disease-specific panels or custom-configured plates. Bio-Rad worked with Thomson Reuters to develop a plate-design strategy that ensures gene assays are the most relevant for gene expression profiling based on differential expression studies and the frequency at which gene targets appear in peer-reviewed literature.
 As two researchers who see the importance of primer validation, both Tennis and Kibschull chose to use Bio-Rad’s collection of PrimePCR products. Not only do the products offer added security, but they eliminate the need to plan and pay for optimization and validation time, letting researchers get back to what really matters: accurate results.
As two researchers who see the importance of primer validation, both Tennis and Kibschull chose to use Bio-Rad’s collection of PrimePCR products. Not only do the products offer added security, but they eliminate the need to plan and pay for optimization and validation time, letting researchers get back to what really matters: accurate results.
Now, with the human embryonic stem cell PrimePCR panel, Kibschull never needs to buy multiple primer pairs and his experiments work every time with 100 percent efficiency and specificity. Tennis is confident that her PrimePCR assays will almost always work on the first try, leaving her worries behind.
When it comes to performing qPCR, researchers might not just be wasting the best part of the roast – they could be cooking the wrong meat altogether. By not validating primers, researchers risk compromising results and integrity. For best results, rather than spending time and money to do it themselves, researchers should choose a solution that they can stake their reputation on.
References
- Marx V. PCR: living life amplified and standardized. Nature Methods. 2013.10, 391–395. doi:10.1038/nmeth.2443. Published April 29, 2013.
- Bustin SA, et al., The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009. 55(4): 611-22. Doi: http://www.ncbi.nlm.nih.gov/pubmed/19246619. Published February 26, 2009.

