Targeted Resequencing: Streamlining NGS for Clinical Research
LISTED UNDER:
With so many new innovations emerging in genomics research, the evolving technology of next generation sequencing (NGS) is becoming an increasingly powerful tool, with researchers now able to simultaneously screen for thousands of disease-linked variants in a single individual. As costs decrease and informatics improve, NGS is now more accessible than ever for use in clinical research, particularly when it comes to understanding complex diseases such as cancer.
Although whole genome sequencing (WGS) provides information on the complete DNA sequence of an individual, the volume of data that this generates makes analysis particularly complex and resource-intensive. Furthermore, for many diseases the relevant genomic region is often already known and genome-wide information is unnecessary. Taking this into account, targeted resequencing, which selects chosen regions of the genome for analysis, offers a highly flexible and economical alternative to WGS.
 An efficient alternative to WGS
An efficient alternative to WGS
Reducing the target size and focusing on the regions most likely to yield relevant data enhances sensitivity through increasing depth of coverage, and therefore the likelihood of new discoveries. Targeted approaches have already had a major impact on disease detection by permitting successful identification of causal mutations for a number of genetic disorders1-5 and cancers.6,7
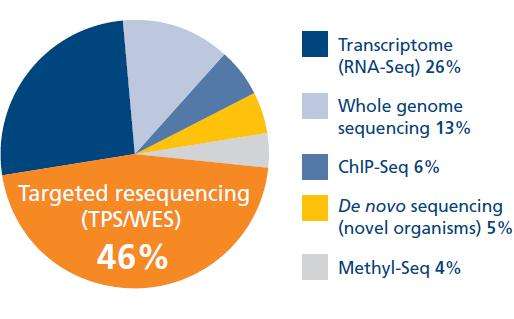
A recent market research survey has even shown that the majority of NGS investigators are now moving towards targeted resequencing techniques where possible (Figure 1).
Where to target?
Targeted resequencing is the term given to the analysis of any defined region of the genome. This comprises whole exome sequencing (WES), which as the name suggests, can be employed to target just the gene-encoding regions and custom panel sequencing, which includes smaller panels focussing on specific regions of the genome. Figure 2 compares the three different approaches of WGS, custom panel sequencing and WES.
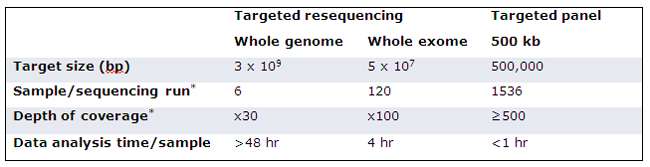
As the exome accounts for only 1.5% of the human genome, and yet includes 85% of all disease-causing mutations,3 WES methods are highly popular for detecting causal variants without compromising the chance of discovering de novo mutations. On the other hand, custom panel sequencing presents a more suitable option if the biological question is more focused. Custom designs can provide information on both intronic and exonic regions that might be associated with a particular disease, or perhaps linked to therapeutic intervention.
Custom panel sequencing also presents options for enhancing the detection of rare variants or variants present in highly heterogeneous samples, as depth of coverage can be further increased when focussing on a smaller target. This approach is ideal for cancer research, as such variants may be missed or under-represented in WES.
Covering all bases—the importance of uniform analysis
One of the greatest challenges remaining for targeted resequencing is obtaining complete and uniform NGS coverage, which in turn depends on unbiased enrichment of all targeted stretches of the genome. The two major enrichment technologies are amplicon-based methods and in-solution capture, and the quality of results generated using either can vary widely.
Amplicon-based strategies employ PCR primers flanking the target, allowing amplification and subsequent library construction. Primer sets can also be designed to encode adaptor sequences for downstream sequencing, and even barcode stretches to enable multiplexing. In-solution methods require additional hands-on time, starting with initial fragmentation of the genome followed by subsequent hybridisation to complementary probes. While amplicon-based methods may be faster with limited hands-on time they are limited by the amount of the genome that can be targeted, as well as variable sequence uniformity due to amplification efficiency. In contrast, although it may be slower, in-solution hybridisation offers superior uniformity and is overall a more efficient strategy, especially for larger target regions (up to an entire exome).
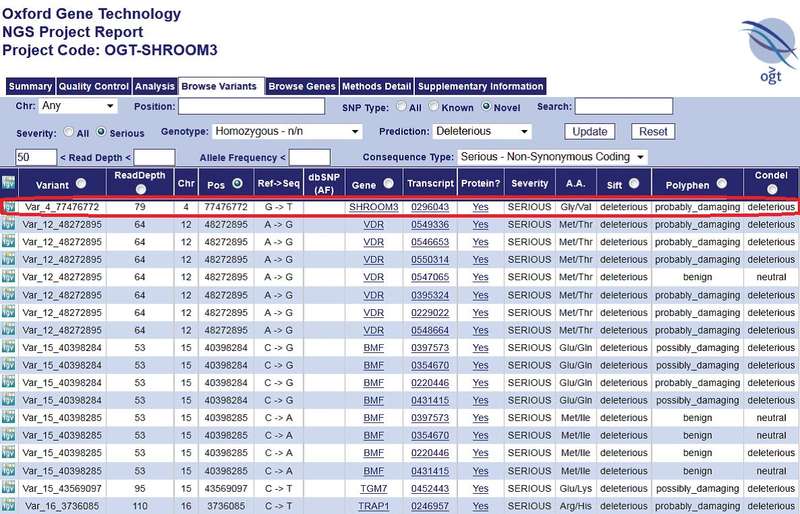 The complexity of both experimental design, not to mention data analysis, make the use of NGS service providers such as Oxford Gene Technology (OGT) with dedicated molecular biology and bioinformatics expertise the ideal option for any sized clinical research programme. Some service providers also deliver results in a user-friendly data analysis report, allowing researchers to rapidly filter through to the results that are relevant to the project, overcoming the common bottleneck of handling the huge data sets produced by NGS applications— with no requirement for an additional bioinformatics resource (Figure 3). Such providers also keep up to date with the latest equipment, allowing researchers to focus purely on interpreting their meaningful results.
The complexity of both experimental design, not to mention data analysis, make the use of NGS service providers such as Oxford Gene Technology (OGT) with dedicated molecular biology and bioinformatics expertise the ideal option for any sized clinical research programme. Some service providers also deliver results in a user-friendly data analysis report, allowing researchers to rapidly filter through to the results that are relevant to the project, overcoming the common bottleneck of handling the huge data sets produced by NGS applications— with no requirement for an additional bioinformatics resource (Figure 3). Such providers also keep up to date with the latest equipment, allowing researchers to focus purely on interpreting their meaningful results.
Disease insights with targeted resequencing
WGS may generate a wealth of data, but for research into genetic disease it is not necessarily better to sequence more of the genome. In fact, many findings from WGS studies may have been found faster and more efficiently using a WES approach, and custom panel sequencing can offer even further flexibility in which specific region to target.
The benefits afforded from targeted resequencing, together with the option to outsource sequencing programmes, is set to transform the use of NGS in the clinical research space and drive exciting new discoveries for genetic disorders.
OGT’s Genefficiency NGS Services are for research use only; not for use in diagnostic procedures.
References
1. Classen, CF. et al (2013) Dissecting the genotype in syndromic intellectual disability using whole exome sequencing in addition to genome-wide copy number analysis. Human Genetics, April 4. [Epub ahead of print]
2. Semler, O. et al (2012) A Mutation in the 5’-UTR of IFITM5 Creates an In-Frame Start Codon and Causes Autosomal-Dominant Osteogenesis Imperfacta Type V with Hyperlastic Callus. The American Journal of Human Genetics 91, 349-357
3. Choi, M. et al (2009) Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proceedings of the National Academy of Sciences of the United States of America 106, 19096-19101
4. Ng, S.B. et al (2010) Exome sequencing identifies the cause of a mendelian disorder. Nature Genetics 42, 30-35
5. Ng, S.B. et al (2009) Targeted capture and massively parallel sequencing of 12 human exomes. Nature 461, 272-276
6. Wei, X. et al (2011) Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nature Genetics 43, 442-446
7. Yan, X.J. et al (2011) Exome Sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nature Genetics 43, 309-315
8. Tariq, M. et al (2011) SHROOM3 is a novel candidate for heterotaxy identified by whole exome sequencing. Genome Biology 12, R91