Co-visualization of mRNA and Protein to Better Link Genotype and Phenotype
LISTED UNDER:
Correlating levels of mRNA and corresponding proteins within cells provides more information linking gene function to phenotype than examining either alone. Separate measurements of RNA and protein merely provide information about two similar but separate cell populations. The ability to study both in individual cells leads to more physiologically relevant data, including information about cell-to-cell heterogeneity within a given sample. In addition, co-visualizing mRNA and intracellular proteins allows for more efficient utilization of sample, which is particularly important when samples are limited, such as when working with primary cells, stem cells or treated cells.
Traditional techniques for RNA analysis, however, do not allow for co-visualization; the methods alter gene expression profiles and can render cells nonviable and unavailable for further protein analysis. Furthermore, expression detected may not reflect the true expression that is seen at the single cell level.
Our studies demonstrate that SmartFlare fluorescent RNA detection probes (EMD Millipore) can address these issues. The new probes, when used in conjunction with immunostaining, allowed GFAP mRNA and intracellular GFAP protein to be assessed simultaneously in individual cells without the need for transfection reagents. Traditional immunostaining was then performed to visualize the protein within the same cell at the same time, allowing researchers to develop a better understanding of the links between genotype and phenotype.
Methods
Cell culture
Primary astrocytes and neurons were prepared from E18 rat hippocampus in a poly-D-lysine-coated 8-well Millicell EZ slide (EMD Millipore). These slides provided easy chamber removal and hydrophobic paint for prevention of cross-contamination between the wells.
RNA detection probes
Cells were grown for 10 days in astrocyte medium, and SmartFlare probes were added for the last 16 hours of incubation. Cells were plated at 120,000 cells per well, in 400 μL medium per well, in each well of an eight-well chamber slide. The cells were incubated overnight at 37°C in media containing 8 μL of the RNA detection probes (diluted 1:20), including Scramble-Cy3 SmartFlare probe and GFAP Rat-Cy3 SmartFlare probe.
These RNA detection probes enter the cell by means of the cell’s natural endocytosis machinery. The technology uses gold nanoparticles bound to sequence-specific oligonucleotide probes; upon binding to the complementary target RNA, a fluorescent signal is released and can be detected on most fluorescence detection platforms. Over time, the probes exit the cell through natural exocytosis, without adverse effects, allowing the cells to be used for further analysis.
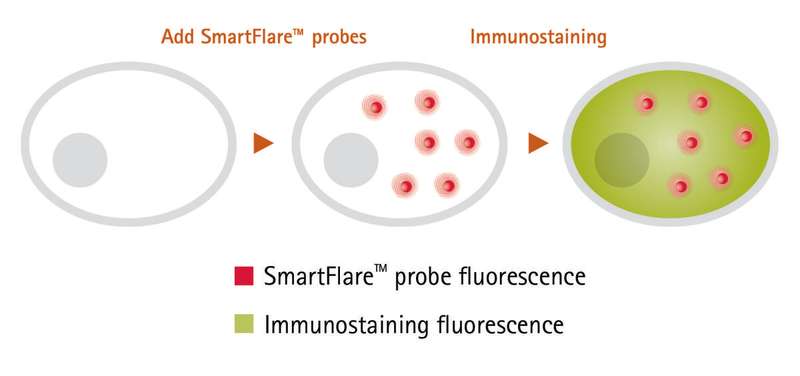 Immunostaining
Immunostaining
Cells may detach easily from the matrix; to minimize disturbance, solution exchanges were performed very gently, with a handheld transfer pipet. Culture medium was removed, leaving 200 μL medium in the well. To avoid disturbing cells, 200 μL of 2X (4%) paraformaldehyde was added to each well and incubated at room temperature for 10 minutes. Fixative was removed, and cells were rinsed once with phosphate-buffered saline (PBS). Cells were permeabilized with 0.1% Triton X-100 surfactant in PBS at room temperature for five minutes, then inspected under the microscope to ensure the cells remained intact. Cells were incubated with antibody solution at room temperature for 30 minutes in the dark. Antibody solution was removed and cells rinsed once with PBS.
Microscopy
The Millicell EZ slide was held down on a flat surface so that the four tabs could be pushed down and broken, enabling easy removal of the wells. The slide was removed from the holder. A coverslip was mounted with mounting fluid and sealed with clear nail polish. Images were obtained using a Nikon C2 confocal fluorescence microscope.
A summary of the methods can be seen in Figure 1.
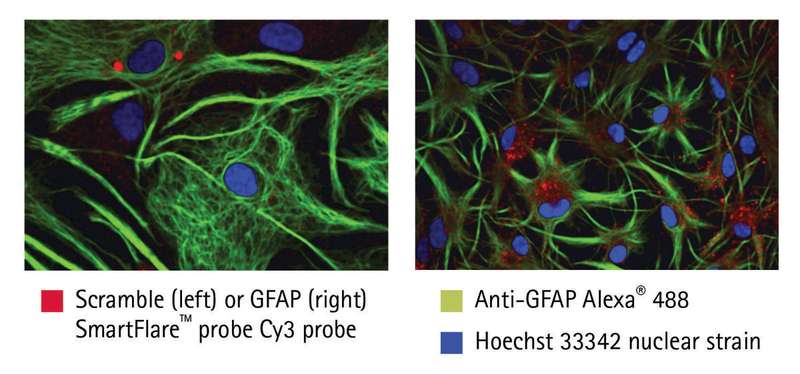 Co-visualization
Co-visualization
RNA and protein were co-visualized in individual, fixed astrocytes using microscopy (Figure 2). Scramble SmartFlare probe (left) showed low background signal, while the GFAP probe (right) revealed punctate fluorescence from GFAP mRNA, an astrocyte marker. In addition, GFAP protein (green) was seen in the cytoplasm of the same astrocytes. In general, cells with higher RNA fluorescence (red) also exhibited higher protein fluorescence (green).
As these results show, RNA detection probes can be used in multiplex with protein detection immunoreagents to study RNA and protein in the same cell, a significant advantage over traditional RNA detection methods. This ability to combine RNA detection with intracellular protein analysis to obtain finer resolution of individual fixed cells by microscopy is critical for scientists looking to analyze mixed populations where cells may not respond uniformly. Armed with this information, researchers can better investigate the links between genotype and phenotype.





