Minimum information for publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines help researchers design qPCR experiments.
Real-time quantitative polymerase chain reaction (qPCR) is a definitive technique for quantifying differences in gene expression levels between samples. However, a lack of consistency in experimental design and reporting combined with inadequate guidelines to review submitted articles with qPCR data greatly increases the potential of reporting statistically insignificant and conflicting results.1 The publication2 and retraction3 of a Science “Breakthrough of the Year 2005” article underlines the issue.
MIQE Guidelines Proposed
To assist the scientific community in producing consistent, high quality qPCR data, the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines were published in 2009. These guidelines aim to increase transparency, promote consistency between laboratories, and therefore, help assure the publication of valid conclusions.4 They also provide a yardstick for the quality assessment of a publication by defining the minimum information required to evaluate PCR data.
The MIQE paper covers a range of qPCR-related topics, including nomenclature, conceptual tips, sample collection and handling, nucleic acid quality control and data analysis. The guidelines also include a checklist that can be used by journal editors to evaluate submitted manuscripts. A simplified version of the guidelines was recently released to help researchers navigate through the design of qPCR experiments in eight simple steps.5
MIQE is modeled after similar guidelines such as MIAME (Minimal Information about a Microarray Experiment) developed several years ago6 and MIAPE (Minimal Information about a Proteomics Experiment).7 All of these initiatives were developed under the umbrella of the MIBBI (Minimum Information for Biological and Biomedical Investigations) standardization body, which has the goal of unifying all of the guidelines for biological and biomedical research.
In addition, the Real-Time PCR Data Markup Language (RDML) has been developed by a consortium to enable the straightforward exchange of qPCR data and related information generated in any lab with a variety of qPCR instruments and third-party data analysis software to assure consistency in submitted data to journals or public data repositories.8 All relevant information about the RDML project is available at www.rdml.org. The recent release of Biogazelle’s product qbase PLUS (www.biogazelle.com/products/qbaseplus) provides users with the only software package that is fully compatible with the MIQE Guidelines. Today, over 4000 researchers worldwide use qBase software for qPCR data management and analysis.
MIQE Guidelines Applied
The MIQE Guideline checklist provides 85 parameters that qPCR studies should be required or recommended to meet before being considered for publication. While a first look at the MIQE guidelines might lead researchers to assume that they will require significant additional effort and slow down the publication process, it is important to note that many of them are discretionary, with only those having the most impact on data quality being mandatory. Adoption of these mandatory guidelines as a first strategy assures that key parameters affecting data quality are being addressed immediately and will have a swift impact on confidence levels in the data and the associated conclusions as recently described.5

click to enlarge |
Table 1. This table summarizes the workflow of a typical RT-qPCR experiment from experimental design to defining control groups, replicates, and experimental conditions to the detailed procedures for sample handling. This ultimately assures that the key steps in RT-qPCR data production lead to high quality, reproducible, and publishable data. (Source: Bio-Rad) |
Experimental Design
Since even minor changes in sample handling and processing can have major changes on the associated mRNA levels, taking the time to define experimental procedures, control groups, type and number of replicates, experimental conditions and sample handling methods within each group is essential to minimize variability (Table 1). Each of these parameters should be carefully recorded prior to conducting gene expression experiments to assure good biological reproducibility for published data.
RNA Extraction
If samples must be collected over a period of time or in too large a number to process immediately, they should be stored in appropriate conditions (frozen at -80ºC and/or in RNA storage solution) until use. They should also be processed in batches of 10-20 to minimize handling time with careful consideration in controlling the sample thaw, RNA extraction processing and associated quality control procedures.
RNA Quality Control
Ensuring the purity (no contaminants) and integrity (not degraded) of the RNA sample is critical. Impurities may lead to inhibition of the reverse transcription and the qPCR reaction giving biased data. The use of partially degraded RNA will lead to inconsistent quantification results and high variability.9 Since sample purity and integrity are not related, both should be assessed to ascertain that the RNA sample meets minimal acceptance criteria for the downstream workflow (Figure 1).10
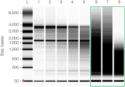
click to enlarge  click to enlarge click to enlarge  click to enlarge click to enlarge  click to enlarge click to enlarge |
Figure 1. Example of qPCR analysis of GAPDH expression performed on varying degrees of degraded liver carcinoma total RNA samples. The profiles show increasing Ct of replicates as the severity of degradation increases. Black traces ar qPCR reactions from undegraded sample (RQI = 9.9), red traces are qPCR reactions from severely degraded sample (RQI = 1.5). (Source: Bio-Rad) |
The purity of RNA is usually measured with a spectrophotometer or NanoDrop based on the OD 260/280 ratio. The traditional method for determining the integrity of RNA is visual inspection of the RNA after electrophoresis on a formaldehyde agarose gel with fluorescent dye. While inexpensive and accessible, this method yields a mostly subjective interpretation of data and requires approximately 200 ng of total RNA which may be difficult if the sample is only available in limited quantities.
A major improvement in RNA integrity analysis came with the introduction of microfluidics-based electrophoresis systems11 such as the Experion automated electrophoresis system from Bio-Rad Laboratories, Inc. Experion combines precise integrity assessment and quantification of RNA in a single step using only nanogram to picogram quantities. In addition to generating a virtual gel, an electropherogram, and calculating the 28S/18S ratio, the Experion software automatically calculates and reports an RNA quality indicator (RQI) value that reflects the integrity of the RNA sample based on several criteria12 of the electropherogram. By assuring consistency in both purity and quality across all RNA samples, variability between biological replicates can be reduced.
Reverse Transcription
Given the prevalence of RNase in the environment, researchers should perform the reverse transcription of total RNA samples to cDNA immediately following the quality control assessment. This will avoid the risk of RNA sample degradation from multiple freeze/thaw cycles before conversion to cDNA. For reverse transcription (RT), the key is to assure consistent and complete coverage of the transcribed genome in the extracted RNA sample. Using the same amount of total RNA and maintaining the same RT reaction time and temperature for all experimental samples is highly recommended to minimize variability between biological replicates. Reverse transcribed RNA can be stored frozen at −20°C or −80°C until use.
Primer and Amplicon Design
Both primer design and careful choice of amplicon (target sequence) are essential to ensure specific and efficient amplification of the products. Target sequences should be unique, 75–150 bp with a GC content between 50–60%, and should not contain secondary structures. It is recommended that primers should have a GC content of 50–60% and a melting temperature of 55–65°C. Long G or C stretches in the primer should be avoided, but it is recommended to have G or C at the end of the primers.
qPCR Validation
A validated qPCR assay is one that has been assessed for the optimal range of primer annealing temperatures, reaction efficiency and specificity using a standard set of samples.5 This will assure that the reaction conditions, buffers, and primers have been optimized and that the cDNA samples are not contaminated with inhibitors of Taq polymerase. Bio-Rad provides qPCR reagents, instruments and software to help researchers achieve consistent qPCR results and adhere to MIQE guidelines.
Bio-Rad has also created a practical Web resource (www.bio-rad.com/genomics/pcrsupport) for qPCR design and validation. The major points in assay validation have been previously described5 and include:
• Determination of annealing temperature, melt curve analysis, gel analysis of amplicon and no template control
• Establishment of a standard curve (to evaluate PCR efficiency)
• Choosing the right qPCR reagents, instrument and analysis program

click to enlarge
|
Figure 2. Experimental replicates. All experiments should be designed with a combination of biological and technical replicates. This illustrates a simple experiment with triplicate biological samples from control and treatment/experimental conditions.For each biological sample, three technical replicates are recommended for the gene of interest as well as for the reference gene(s). This results in a total of at least 36 samples plus the duplicate no template (NTC) for a total of >40 wells. (Source: Bio-Rad) |
Data Analysis-Experimental Reproducibility
There are two sources of variability in a gene expression experiment that may affect the results: biological variability (which is due to inherent differences in gene expression levels between individual organisms, tissues or cell culture samples) and technical variability (which is due to the experimental process itself and is typically associated with pipetting, poorly calibrated pipettes, RNA extraction and downstream processing techniques). To mitigate the effect of biological and technical variability, it is generally accepted that at least three biological and two technical replicates per biological replicate be performed for each experiment (Figure 2).5 If the experiment compares gene expression levels between control and treated samples, the three biological replicates should be samples that were treated in separate and independent experiments.
Conclusion
qPCR is a powerful, enabling and widely adopted technology that can greatly advance scientific research. However, the combination of uninformed use of prepackaged kits and software with a lack of rigorous and standardized methods can lead to a widespread misinterpretation of data and consequent publication of erroneous conclusions. Following the guidelines presented here and the MIQE checklist will improve the reliability of the published results and their interpretation.
About the authors
Sean Taylor, PhD, is a Senior Field Application Specialist at Bio-Rad Laboratories in Canada.
Mary Grace Brubacher, MSc, is a marketing manager at Bio-Rad Laboratories in Hercules, California.
References
1. Garson JA, Huggett JF, Bustin SA, Pfaffl MW, Benes V, Vandesompele J, Shipley GL. Unreliable real-time PCR analysis of human endogenous retrovirus-W (HERV-W) RNA expression and DNA copy number in multiple sclerosis. AIDS Res Hum Retroviruses. Forthcoming 2009.
2. Huang T, Bohlenius H, Eriksson S, Parcy F, Nilsson O. The mRNA of the Arabidopsis gene FT moves from leaf to shoot apex and induces flowering. Science 2005;309:1694–6.
3. Bohlenius H, Eriksson S, Parcy F, Nilsson O. Retraction. Science 2007;316:367.
4. S. Bustin et al. The MIQE Guidelines: Minimum Information for Publication of Quantitiative Real-Time PCR Experiments, Clin Chem. 55, 611-22 (2009).
5. S. Taylor et al. A Practical Approach to RT-qPCR — Publishing Data That Conform to the MIQE Guidelines Bio-Rad Bulletin #5859 (2009).
6. A. Brazma et al. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data, Nat Genet. 29, 365–71 (2001).
7. C.F Taylor et al. The minimum information about a proteomics experiment (MIAPE). Nat Biotechnol. 25, 887-893 (2007).
8. S. Lefever et al. RDML: structured language and reporting guidelines for real-time quantitative 9 PCR data, Nucleic Acids Res. 37, 2065-2069 (2009).
9. Fleige and Pfaffl. RNA integrity and the effect on the real-time qRT-PCR performance, Mol Aspects Med. 27, 126-139 (2006).
10. J. Gingrich et al. Effect of RNA Degradation on Data Quality in Quantitative PCR and Microarray Experiments. BioRad Bulletin #5452B.
11. Imbeaud et al. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces, Nucleic Acids Res. 33, e56 (2005).
12. Denisov et al. Development and validation of RQI: An RNA quality indicator for the Experion automated electrophoresis system, Bio-Rad Bulletin #5761 (2008).