A Guide to Targeted NGS: Generating Accurate Data for Personalized Medicine
As personalized medicine is integrated into mainstream medical treatment, sequencing clinically relevant genes using the latest next generation sequencing (NGS) technologies will prove instrumental in guiding clinicians towards informed treatment decisions.
For many, target capture, which focuses on selected genomic regions, can deliver a more practical alternative to whole genome sequencing, as it significantly decreases cost and the amount of data analysis required. However, this technique must be carefully optimized to ensure the return of accurate data. Geneseeq Technology is a cancer diagnostics company that provides treatment solutions based on the comprehensive, systematic analysis of patients’ genomic mutations using primarily NGS techniques. In this article, Geneseeq scientists share their valuable insight on how to achieve successful target capture, from choosing the technology and reagents to optimizing hybridization conditions.
1. Which target capture technology?
Methods for isolating cancer-related regions from the entire genome for sequencing have traditionally been challenging. The selection of an appropriate approach should therefore take into consideration:
- Sample type— Sufficient yield and quality of input DNA are required from the sample, necessitating optimized and often time-consuming sample preparation from clinical specimens.
- Sensitivity— Maximizing enrichment from the gDNA library requires efficient hybridization and post-capture PCR amplification.
- Uniformity of coverage— Enrichment can be affected by the DNA sequence: GC content and repeats can lead to uneven coverage and, hence, inaccurate NGS data. Targets must, therefore, be carefully chosen.
- Specificity— Reducing the presence of off-target fragments in the sequencing library is vital for assay specificity. Increased specificity can be achieved through optimized hybridization conditions and use of blocking oligos.
Use of oligonucleotide probes is becoming routine for isolate specific genomic regions via in-solution hybridization. The quality of the oligos used for hybridization is crucial for obtaining reliable data. Individually synthesized probes ensure that each desired probe is present in the probe pool or panel. In this format, these are particularly useful for creating custom panels that can be optimized, expanded, and combined as necessary.
Geneseeq scientists found that target capture panels comprising their choice of probes combined in a 1:1 molar ratio delivered ~40% higher enrichment efficiency than other commercially available reagents. Moreover, having individually synthesized probes provided Geneseeq with the flexibility to adjust their panels by adding or removing individual targets or genes as necessary.
2. Optimizing your assay for sample type
DNA can be isolated from a variety of sample types, including fresh and frozen tissue or blood. For effective clinical application, the chosen target capture assay must be compatible with all sample types. Most cancer specimens are formalin-fixed and paraffin-embedded (FFPE), which causes varying degrees of DNA damage. Typically, FFPE samples are notorious for yielding nucleic acid of low quantity and poor quality that is unsatisfactory for library construction and PCR amplification. Optimizing target capture from these samples, therefore, also requires certain considerations:
- Pre-enrichment— Choose appropriate methods of DNA extraction and fragmentation, in addition to library construction.
- Enrichment— When multiplexing, remember that PCR efficiency varies with sample type and DNA quality. To achieve even, optimum coverage for every sample, it is important to adjust library input ratios.
- Post-capture amplification— To reduce inherent PCR duplication and bias in downstream NGS, maximize recovery of the enriched library and minimize PCR cycling.
When working with FFPE samples, Geneseeq scientists found that xylene works best for dissolving paraffin, while heating samples at 90°C for 1 hour after proteinase K digestion reverses some of the formaldehyde modification of nucleic acids. Commercial DNA repair kits also help to improve library quality by repairing a broad range of damage, including mutations and PCR-blocking modifications.
3. Hybridization buffers and post-capture PCR
Adopting an on-bead method for post-capture PCR amplification increased both speed and DNA yield (Figure 1). However, initial experiments revealed saline sodium citrate in the post-capture wash buffer as a potent PCR inhibitor. The solution was to use the Rapid Protocol for DNA Probe Hybridization and Target Capture from Integrated DNA Technologies (IDT), which incorporates a 4 hour hybridization at 47°C using a commercial hybridization and wash kit. Raising the hybridization to 65°C further increased enrichment.
4. Reducing background
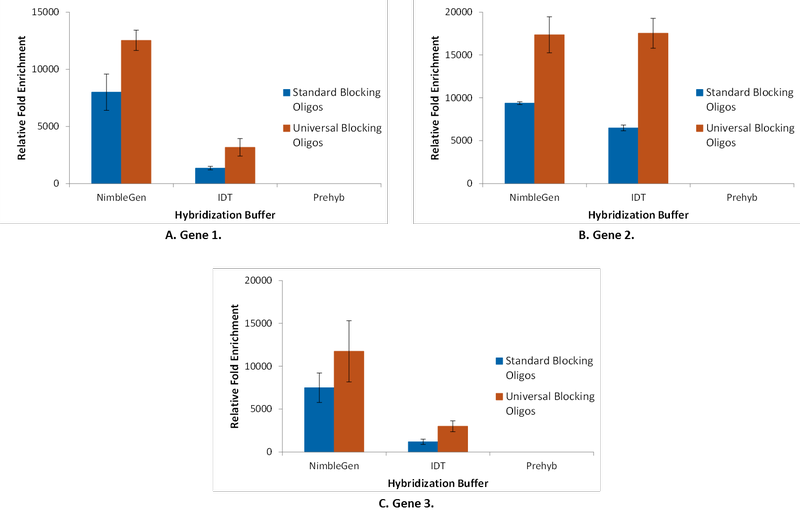
Blocking oligos added during the hybridization reaction reduced the off-target rate and improved target capture specificity. Geneseeq recommends a universal blocking oligo based on a single oligo sequence that blocks multiple barcoded adapters. The researchers found that, compared to standard blocking oligos, these oligos were more efficient at reducing background, leading to enhanced enrichment (Figure 2).
In fact, while negative controls (ACTB [β-actin] and GAPDH) were detected within the gDNA library, no trace of these controls remained after enrichment in the presence of universal blocking oligos. This finding demonstrates that an optimized assay can effectively deplete off-target sequences.
Translating research into clinical diagnostics
Geneseeq has tested >300 patients in the last year and is continuing with longitudinal studies of some of these patients. Accurate NGS data is crucial for this work, and with targeted sequencing this can only be achieved with a sensitive, robust target capture assay. Performing an in-depth protocol optimization experiment, the researchers found that their optimized target capture protocol achieved >80% on-target rate with high uniformity of coverage for different genes. Through accumulating diagnosis and treatment information from a large number of patients, Geneseeq is generating data to provide statistical significance to its tests for application in clinical practice.



_itok-ikct0yc3.jpg)

