Using Surface Plasmon Resonance with Phage Display Libraries
 Surface Plasmon Resonance (SPR) and phage display are both techniques that are well suited to high-throughput environments. In particular, phage display has found utility in antibody production. Phage display can be used for recombinant antibody isolation. Producing antibodies via phage display is seen as a step forward when compared to traditional methods. Phage display can also be used for the study of other types of samples including peptides. SPR has been explored as a means of substantially reducing the time and number of steps required for phage antibody isolation, while simultaneously providing kinetics and affinity information that are the hallmarks of the SPR technique.
Surface Plasmon Resonance (SPR) and phage display are both techniques that are well suited to high-throughput environments. In particular, phage display has found utility in antibody production. Phage display can be used for recombinant antibody isolation. Producing antibodies via phage display is seen as a step forward when compared to traditional methods. Phage display can also be used for the study of other types of samples including peptides. SPR has been explored as a means of substantially reducing the time and number of steps required for phage antibody isolation, while simultaneously providing kinetics and affinity information that are the hallmarks of the SPR technique.
Traditional antibody production
Antibodies have traditionally been made by injecting an antigen into a mouse. This leads to the production of an immune response in the mouse. The mouse is then dosed with the same antigen over the course of several weeks until the level of antibodies in the serum is high enough. The animal is then euthanized. For production of monoclonal antibodies (those produced from a single clone of B cells), immune cells from the mouse spleen (which have a limited lifetime) are then removed and fused with cancer cells (which have essentially an unlimited lifetime) to produce a hybridoma. Myeloma cells have typically been used as the source for the cancer cells for this process. This is followed by either injection into an animal, producing ascites, or by cell culturing, to produce a continuous supply of antibodies. The process is quite time consuming, taking months to obtain a supply of specific monoclonal antibody. Production of polyclonal antibodies (those produced from multiple clones of B cells) is faster, but still tales about 4-8 weeks.
Recombinant antibodies and phage display libraries
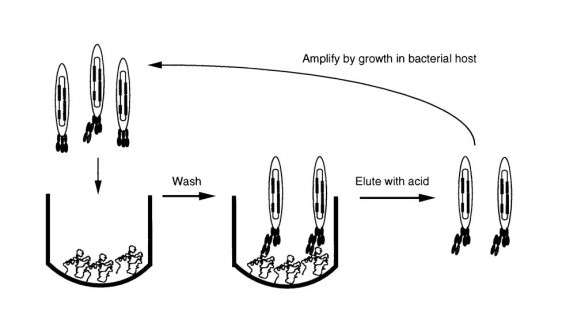
Development of antibodies has moved towards recombinant methods and away from hybridoma production. This eliminates most of the issues related to traditional antibody production. Among the improvements, no animals are needed. Recombinant technology eliminates all the animal immunization steps, thus eliminating the traditional limitations imposed by needing to induce an immune response in the laboratory mouse (or other animal). There are a number of backbones to use for production of recombinant antibodies including ribosomes, cells, and DNA. Another way to do it is to use a bacteriophage (or “phage”) as the backbone for construction of antibody libraries. Filamentous phages of E. coli can be used. Researchers often pool blood samples to use in production of phage libraries. A collection of phage is called a “phage display library”. Each phage particle displays a particular antibody and antibody genes are fused to phage genes. Each “library” can contain ~1x108 to 1x1011 clones. Once the library is created, “panning” can be carried out. The phage is successively put in contact with antigen (for example, in wells on an ELISA plate) to selectively separate out antibodies. The “panning” is repeated until purity becomes high enough to use. Using this technique, production of antibodies can take only about one week - a big improvement in time compared to traditional methods.
 Using SPR with phage display libraries
Using SPR with phage display libraries
Surface Plasmon Resonance (SPR) has traditionally been used to study biomolecular interactions. Typically, one half of a binding pair (the target) is coupled to the surface of either a carboxymethyl dextran sensor chip or a sensor chip with a self-assembled monolayer (SAM) on the surface. When the other half of the binding pair (the analyte) is flowed over the surface, the binding interaction is followed. Kinetics are obtained by injecting a series of concentrations of the analyte over the target and fitting the curves globally to get on (association) and off (dissociation) rates as well as the equilibrium dissociation constant (KD). If the interaction does not dissociate all the way back to baseline, then a regeneration step is used after each sample injection. The regeneration solution is chosen such that it removes all the bound analyte but does not denature the target on the surface. In this case, a specific antigen can be amine coupled to a SPR sensor chip. While ELISA has traditionally been used to rank affinity, SPR has certain advantages. A big advantage of SPR versus ELISA is that a much lower amount of target is needed for SPR experiments. Also, with SPR, almost any target can be coupled to the sensor chip surface. SPR can also be used as a faster way to obtain pure higher affinity antibodies from phage display libraries – SPR can essentially be used to do the “panning”. The filamentous phage can be injected over a target immobilized on the sensor chip surface and only specific antibodies to the immobilized antigen or other target will bind. These antibodies can then be removed via regeneration and collected for further use (testing or further selection). This process can be repeated using a number of different targets to separate out the specific antibodies and to further accurately determine their affinity.
In summary
Improvements in antibody production have occurred in recent years, particularly with the move to recombinant technology. Some interesting improvements have already been realized for antibody production by using SPR and phage display together. Others may be possible. For instance, SPR parameters can be adjusted to provide improved separation and affinity ranking when running phage– changing things like flow rate, temperature, using gradient elution, etc. are interesting topics for future research.